 Demo screenshot
Demo screenshotAbstract
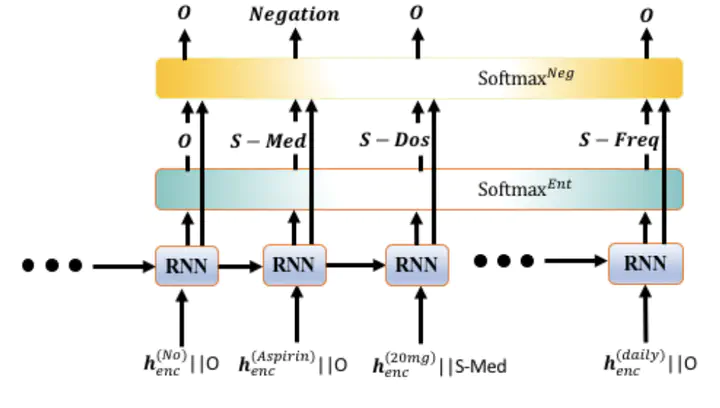
Negative medical findings are prevalent in clinical reports, yet discriminating them from positive findings remains a challenging task for in-formation extraction. Most of the existing systems treat this task as a pipeline of two separate tasks, i.e., named entity recognition (NER)and rule-based negation detection. We consider this as a multi-task problem and present a novel end-to-end neural model to jointly extract entities and negations. We extend a standard hierarchical encoder-decoder NER model and first adopt a shared encoder followed by separate decoders for the two tasks. This architecture performs considerably better than the previous rule-based and machine learning-based systems. To overcome the problem of increased parameter size especially for low-resource settings, we propose the Conditional Softmax Shared Decoder architecture which achieves state-of-art results for NER and negation detection on the 2010 i2b2/VA challenge dataset and a proprietary de-identified clinical dataset.
Supplementary notes can be added here, including code, math, and images.