Mohammed Khalilia (محمد عبد الستار قاسم)
Senior Applied Scientist | Adjunct Professor
Qualtrics
Birzeit University
Biography
Mohammed Khalilia (محمد عبد الستار قاسم) is a researcher, computer scientist, programmer and data scientist who earned his PhD in computer science from the University of Missouri. He joined Georgia Tech Computational Science and Enginerring school and Emory University as Postdoctoral Fellow to work on predictive modeling in healthcare. His research included predictive modeling, relational cluster analysis, health and nursing informatics. He then worked for Amazon Web Services (AWS) and Amazon Studios for about five years in the areas of natural language processing (NLP), speech synthesis and some computer vision. In 2018, he helped launch Comprehend Medical, an Amazon web service for medical NLP.
Currently, he is a Senior Applied Scientist at Qualtrics working on conversational machine learning and active learning. He is also an adjunct professor at Birzeit University teaching NLP related courses for doctoral students.
- Machine learning
- Natural Language Processing
- Biomedical/health informatics
- Predicitive modeling
PhD in Computer Science, 2014
University of Missouri
BS in Computer Science, 2006
University of Missouri
Featured Publications

This paper presents Wojood, a corpus for Arabic nested Named Entity Recognition (NER). Nested entities occur when one entity mention is embedded inside another entity mention. Wojood consists of about 550K Modern Standard Arabic (MSA) and dialect tokens that are manually annotated with 21 entity types including person, organization, location, event and date. More importantly, the corpus is annotated with nested entities instead of the more common flat annotations. The data contains about 75K entities and 22.5% of which are nested. The inter-annotator evaluation of the corpus demonstrated a strong agreement with Cohen’s Kappa of 0.979 and an F1-score of 0.976. To validate our data, we used the corpus to train a nested NER model based on multi-task learning using the pre-trained AraBERT (Arabic BERT). The model achieved an overall micro F1-score of 0.884. Our corpus, the annotation guidelines, the source code and the pre-trained model are publicly available.
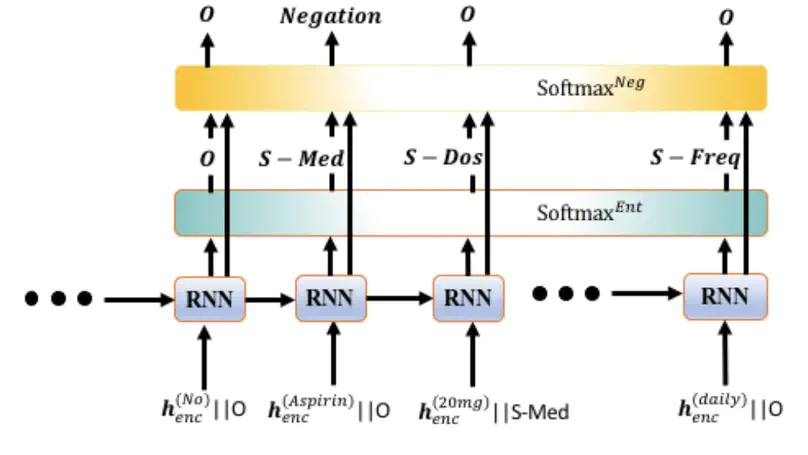
Negative medical findings are prevalent in clinical reports, yet discriminating them from positive findings remains a challenging task for in-formation extraction. Most of the existing systems treat this task as a pipeline of two separate tasks, i.e., named entity recognition (NER)and rule-based negation detection. We consider this as a multi-task problem and present a novel end-to-end neural model to jointly extract entities and negations. We extend a standard hierarchical encoder-decoder NER model and first adopt a shared encoder followed by separate decoders for the two tasks. This architecture performs considerably better than the previous rule-based and machine learning-based systems. To overcome the problem of increased parameter size especially for low-resource settings, we propose the Conditional Softmax Shared Decoder architecture which achieves state-of-art results for NER and negation detection on the 2010 i2b2/VA challenge dataset and a proprietary de-identified clinical dataset.
Recent Publications
Contact
- firstname.lastname@gmail.com
- 1201 2nd Avenue, Seattle, Washington 98101
